NOTE: After creating this blog post, I created this Biostars discussion. I think this is a little shorter and perhaps a better format for discussion. So, you can want to consider looking at that discussion instead of (or in addition to) this post. Thank you very much for your interest.
As also mentioned in this other post, my African ancestry (whether that is what most people would consider to be African, or ancestors that migrated out of Africa relatively more recently) should come from my father's side.
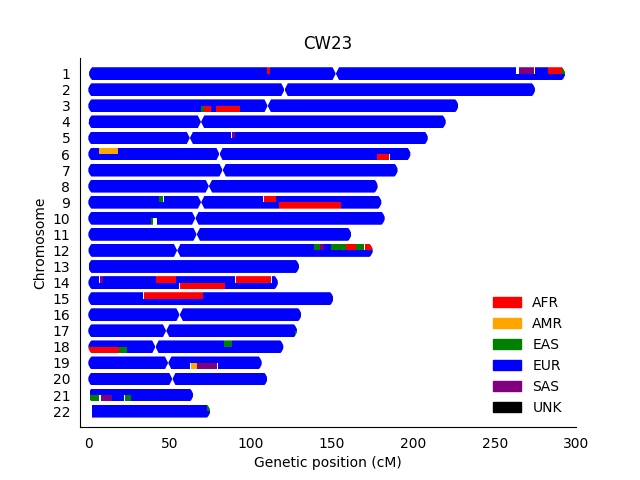
While upstream phasing by SHAPEIT can also be a factor, I did some RFMix re-analysis with various data types, including the result below:
Assuming that each row represents a chromosome that I inherited from each parent, the most clear problem is that the African (AFR red ancestry) should really only be on one copy of chromosome 14 (and I assume the adjacent European EUR segments are not precise, and there should really be one larger segment).
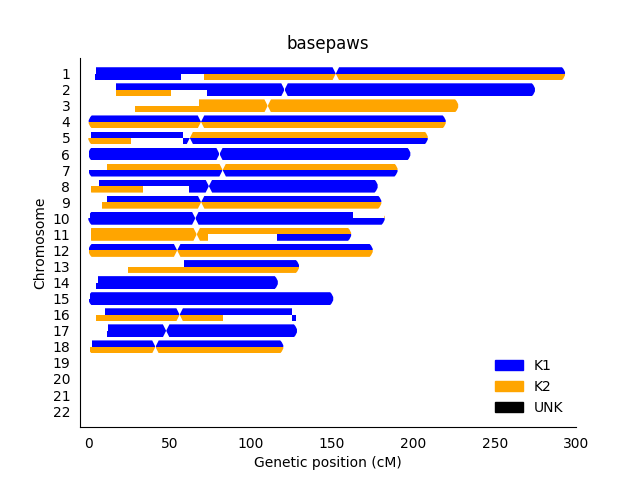
While I am not entirely certain about the underlying method, there is also an example where I think visual inspection can be useful for a result from basepaws. While my notes are messy (and sometimes incomplete), you can look here for more information (if desired). I purchased ~15x Whole Genome Sequencing for $1000 (to get raw data), rather than the more typical $95 for low coverage Whole Genome Sequencing (lcWGS) and Amplicon-Seq for health markers.
So, I don't exactly have my own basepaws report, but I think there is a fairly new version of broad ancestry assignments (via chromosome painting) that can be viewed on page 3 of this PDF (for another cat). In terms of separate images that I can find on-line, this blog post with an earlier report (again, for another cat) has ancestry painting, but it doesn't have the same problem. Likewise, the chromosome painting plot on this blog post doesn't have the same issue.
So, I will just verbally say that the cat chromosome painting on on page 3 of this PDF looks off in that the broad ancestry assignments seem to be the same on both copies of each chromosome. To be fair, they aren't always the same (which is good - I think the ancestry for most cats should probably be relatively independent for each chromosome copy).
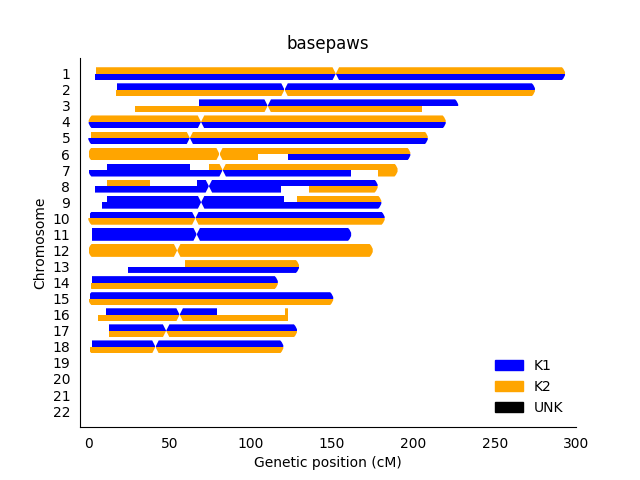
However, in terms of giving advice for troubleshooting, I can also show you my attempted RFMix analysis (which clearly has problems, and I wouldn't recommend for returning as a result for anybody else).
Now, the chromosome copies do show more independent ancestry (per chromosome copy), but the results are not reproducible. However, in that particular context (performing re-analysis of my cat's data), I thought ADMIXURE and PCA (using public reference samples) did have reasonable results. So, I think there were other strategies (which I would probably consider "simpler" strategies that I thought did give reasonable results), which is good and important. While there may be a problem in the assumption of use for some more specific breeds (such a single markers for the Scottish Fold or Sphynx), my point is that I consider the ADMIXTURE and PCA results to be OK for the broader ancestry (meaning I think there is some sort of robust ancestry result that can be provided for cats).
Nevertheless, the overall goal was to be able to visually identify likely problems with inheritance (and/or limitations to precision for ancestry results), and I think the above plots are OK for that.
That said, even this troubleshooting should probably be thought of as "hypothesis generation." In other words, if your first assumption when seeing results like I have shown above is not "Something looks like it could / likely is wrong," then I think this is helpful in terms of needing to critically assess genomics results. However, it is also important that you then try to think of ways to identify the more specific problem. While phasing may be an issue with the human 23andMe results and the more limited number of probes / markers for the public reference samples is my expected problem with the basepaws cat RFMix analysis, you generally gain confidence in a result when you keep trying find a problem (and you keep finding valid explanations for the results). So, in some ways, it may be best to call my critique a "hypothesis." For example, saying I couldn't get reasonable RFMix results for my cat analysis (and I should also admit that there could also be some sort of bug that I haven't been able to find) is not the same of saying some sort of chromosome painting analysis is not possible in the future (as long as the underlying biological assumption is valid).
Change Log:
9/16/2019 - public post date
2/6/2020 - add Biostars discussion link





No comments:
Post a Comment