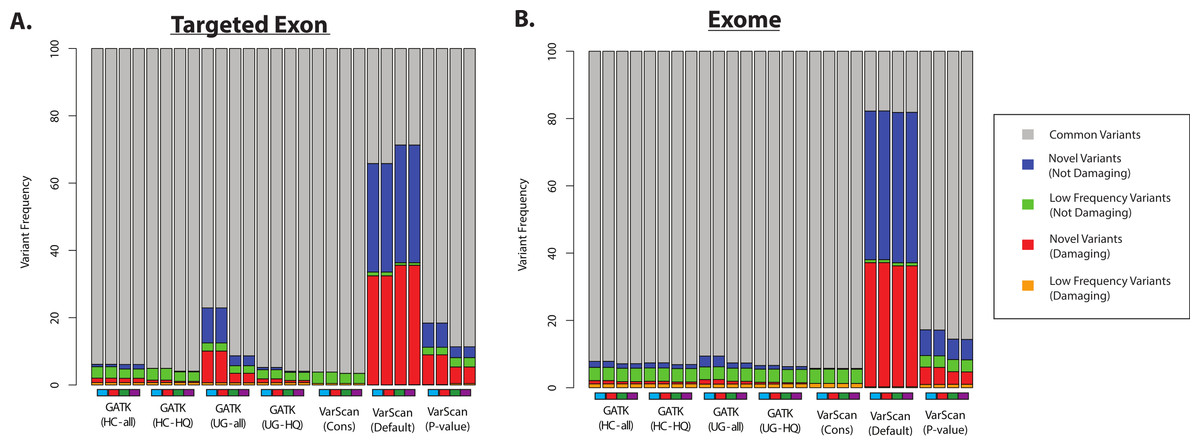
I thought the description of a species that was in between a mouse and a monkey was interesting, but I am not convinced about the widespread use of the mouse lemur as a model organism. I'm also not sure about the evidence for the mouse lemur being the 2nd most abundant primate (after people); for example, one of the "least-concern" mouse lemur species had a unknown population size on one website (and the Wikipedia link lists a gibbon as being 2nd, with an order of magnitude difference in population size). However, if there are ways to learn about possible human treatments through the experiences with cost effective ways to help the mouse lemur population, then I think that would be extremely interesting.
I believe I first heard about human treatments being inspired by animal treatments when I was reading the article "Cancer Clues from Pet Dogs," in the 2018 special issue of Scientific American on "The Science of Dogs and Cats." I don't believe I saw specific citations for the points that I found most interesting, but I'll summarize some points below (with some papers that I found by Googling):
- My understanding is that piroxicam treating bladder cancer in dogs influenced the use of celecoxib (Celebrex) in humans.
- Perhaps this relates to what is described in Knapp et al. 2016?
- The VCA link above mentions the use being off-label, so I am guessing there is even more of a backstory.
- The article also mentions Stephen Withrow playing a role in optimizing methods to avoid limb amputation of osteosarcoma (in dogs, but now used in people).
- I also found a description of that procedure being applied to cats on-line.
- The article mentions intranasal delivery of IL-2 contributing to inhaled IL-2 therapy for lung metastases in people
- I didn't see a top Google hit for exactly what I was looking for, but I a similar description in this Google Books document.
- David Waters (one of the authors) describes using CT scans in dogs to test whether signal is for a true tumor (or a false positive), with the expectation that should also help improve the accuracy in the methods for people
- I think this study by Waters et al. 1998 is a relevant citation, although I am not sure if there is a paper to show translation into human methods
- I'm also not entirely certain if we are talking about the dogs as patients (those are the examples that I am trying to list), versus the dogs as subjects (which would be more along the lines of a "model organism" or "animal model")
Again, I'm not sure what are the primarily references for these claims, where I would ideally want to see validation from independent labs. Admittedly, that is for biological / medical concepts, and you can't really prove inspiration in the same sense (there are limits in the chronological order, when claiming "procedure X in dogs enabled the similar or same procedure X in people," although I've certainly seen examples where the official publication date of peer reviewed publications didn't actually match the order of discovery). Nevertheless, please let me know if you can help me fill in some of the details!
More broadly, in "Reason for Hope" (in the chapter "On the Road to Damascus"), Jane Goodall describes somebody whose's daughter had a heart problem and was told "her daughter was only alive because of experimental work on dogs," and that individual approached Jane in a very negative manner. Dr. Goodall then waited for the individual to calm down, and explained that her mother had a big valve in her heart (even thinking through the process to point out "[it] was from a commercially slaughtered hog, but the procedure had been worked out with pigs in a laboratory"). She then said "I just feel terribly grateful to the pig that saved my mother's life...So I want to do all I can to improve conditions for pigs - in the labs and on the farms. Don't you feel grateful to the dogs who saved your daughter?" I think this is a good mindset. From my end, I also think that treating animals more like people might even help with successfully translating discoveries to humans.
I also think that there are fewer experiments on cats and dogs now (most mammalian experiments are in mice), but I don't believe those numbers have dropped to zero. That said, if you start asking people what evidence they want to see before an experiment is performed on their own pet, then that may also shift the way some people think about animal research.
Also, the video mentions creation of knock-out mutants (for a model organism, like a mouse), and this is different than the animal therapies that I describe above. However, I think communication between those representing animal rights and those conducting animal experiments is important. For example, you need mutual understanding and open communications to have discussions about whether an experiment is necessary or (in the case of the genetic modification, like a gene knock-out) how to troubleshoot if that modification was successfully and retained over generations. This might even be relevant to discussions of safety for genetic editing in people.
Finally, pets may not always have health insurance (and wildlife definitely doesn't have insurance). For example, I'm going out on a limb on this one, but maybe this could even help with some of the details of directly offering generics through non-profits. So, if we can find cost effective ways to make discoveries and continue to provide treatment at a reasonable cost (while respecting those who contributed to that body of knowledge), then I think that could be quite interesting and important.
Change Log:
6/22/2019 - public post date
-- I also want to acknowledge that I had a Facebook post about the mouse lemur article, and that is why I expanded my thoughts for this post.
11/3/2019 - add "speculative opinion" tag