NOTE (2/24/2020): Nebula is currently offering 30x sequencing. So, my concerns about the low coverage Whole Genome Sequencing (lcWGS) at ~0.5x are probably less relevant for that particular company. However, if you get lcWGS from another company, then this information is probably relevant.
Concerns about Specific Variants
While I very much support providing FASTQ, BAM and VCF data, one of my concerns about the Nebula results was the use of low-coverage sequencing.
So, one of the first things that I did was visualize the alignments for some of my more confidently understood variants from previous data (using IGV).
For the two alignments below, the Genos Exome is the top alignment, the Nebula low-coverage alignment is in the middle, and the Veritas Whole Genome Sequencing (WGS, regular-coverage) is at the bottom.
My cystic fibrosis variant (rs121908769):
My APOE Alzhiemer's risk variant (rs429358, Nebula alignment in middle, variant is red-blue bar in the right-most exon):
For APOE, I zoomed out from the screenshot so that you could get a better perspective of the error rate per-read at other positions around the gene.
You could see my cystic fibrosis variant in the 1 read covered at that position, but you can't see any reads with the APOE variant. My concern about the use of low-coverage sequencing is due to imputation (at least for traits). Even though this APOE variant is somewhat common (I believe ~15% of the population), the imputation failed to identify me as having that variant. You see that from the .vcf files
My APOE Alzhiemer's risk variant:
19 45411941 rs429358 T C . PASS . GT:RC:AC:GP:DS 0/0:0:0:0.923102,0.0768962,1.71523e-06:0.0768996
As described in the gVCF header:
GT = Genotype
RC = Count of Reads with Ref Allele
AC = Count of reads with Alt Allele
GP = Genotype Probability: Pr(0/0), Pr(1/0), Pr(1/1)
DS = Estimated Alternate Allele Dosage
Update (8/5): Nebula support got in touch with me and explained that the blog post is in reference to Nebula Research Library (rather than the "Your Traits" section). I canceled my subscription, I am still able to confirm that I see this under the "Library" section (rather than "Traits," "Ancestry," or "Microbiome").
Likewise, there was no delTT variant in the VCF, so my cystic fibrosis carrier status would also be a false negative (if that was used in the report), even though you could actually see that deletion in the 1 read aligned at that position (because 1 read wasn't sufficient to have confidence in that variant).
Overall Variant Concordance
I can also use my VCF_recovery.pl script to compare recovery of my Veritas WGS variants in my Nebula gVCF.
If you compare SNPs, then the accuracy is noticeably lower than GATK (and even lower than DeepVariant):
3,071,596 / 3,419,611 (89.8%) full SNP recovery
3,184,641 / 3,419,611 (93.1%) partial SNP recovery
The indels are harder to compare (becuase of the freebayes indel format). So, in the interests of fairness, I am omiting them here (as I did for comparing the provided Genos Exome versus Veritas WGS variants). However, instead of comparing the provided Veritas WGS .vcf file, I can try comparing the BWA-MEM re-aligned GATK Veritas WGS .vcf (which also had higher concordance between my Exome and WGS datasets):
3,133,635 / 3,419,611 (91.6%) full SNP recovery
3,248,277 / 3,419,611 (95.0%) partial SNP recovery
164,140 / 217,959 (75.3%) full insertion recovery
180,736 / 217,959 (82.9%) partial insertion recovery
190,452 / 266,479 (71.5%) full deletion recovery
213,131 / 266,479 (80.0%) partial deletion recoveryThe GATK recovery is a little better. However, it is very important to emphasize that the gVCF variants do not have 99% accuracy (even for average accuracy, or even for SNPs). I think whatever benchmark was used for that calculation was probably over-fit on some training data. To be fair, I think the average SNP chip concordance (with higher coverage WGS data) is also lower than some people might expect, but it is definitely higher than this lcWGS data.
You can also show similar results with precisonFDA (using the BWA-MEM realigned GATK gVCF, which we expect to have better concordance than the provided Veritas gVCF).
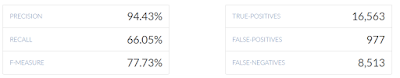
For example, the overall file shows noticably low recall when comparing the Nebula imputed gVCF versus the Veritas WGS BWA-MEM re-aligned gVCF:
and, to be more fair for the Exome versus WGS comparison in the blog post, the trend is similar within RefSeq CDS regions:
The screenshots are smaller than in the blog post because there was no precision-recall plot for the Imputed Nebula gVCF comparisons.
So, I disagree with the use of low-coverage sequencing for traits, and I would respectfully consider removing this section (or only made available to those with higher-coverage sequencing).
When I was trying to upload my raw data to my Personal Genome Project page, I noticed that they had an option called "genetic data - Gencove low pass (e.g. Nebula Genomics)". This makes me think discouraging low-coverage sequencing is something that needs to be done more broadly (at least for health traits).
Concerns abut Nebula Library Results
My concerns for the previous sections are probably solved when using the higher coverage sequencing data. So, unless you were an earlier customer and had the lower coverage sequencing data, you probably don't have to be extra careful about possibly overestimated accuracy in your genotype imputations.
However, there is one thing that I think could still be a problem for customers with higher coverage sequencing data (if the reports are the same). The concept is similar to my concern about the basepaws breed index (described in this blog post) and/or other Polygenic Risk Scores that I have collected for myself, but I think I can explain my concern with the top 3 percentile results that I received from Nebula:
As you can see from this link, the percentile above was calculated using 13 SNPs. Seven of the thirteen SNPs are on chromosome 6, and 5/7 of those variants didn't have alignments against the main reference chrososome (for hg19) in my higher coverage Veritas Whole Genome Sequencing data. Nebula predicted 3-4 of those 7 chromosome 6 variants to be homozygous variants, but I am not sure if these are correct or not (and the nucleotide for rs3763312 was different from the variants in dbSNP). There were a pair of variants on chromosome 10 where I was predicted to be heterozgyous at both sites. For the remaining 6 non-chr6 variants, I had imputed genotypes for 2 heterozygous variants, 2 homozygous non-reference variants, and 2 homozygous reference variants (and they matched my higher coverage WGS data).
I am only 34, but I definitely don't have hair that looks like the Google images for this disease. So, I don't know the expected age of onset, but I think I might never get this condition (even though Nebula says that I am at the 100th percentile).
As you can see from this link, the percentile above was calculated using 15 SNPs. 12 of those SNPs had at least 1 variant from the reference genome and 11/12 of those variants matched by Vertias WGS variants. The discordant variant was rs6910071, which was homozygous for the variant allele on the Nebula lcWGS imputed variants. So, this could have been consistent with 90% overall accuracy, but I didn't have coverage for either dataset at this position (so, this isn't the same as using a gVCF to make a homozygous reference genotype call).
I have osteoarthritis in my lower back, but I don't believe that I (currently) have rheumatoid arthritis.
While I could believe that I am at increased risk, it is important to note that the summary only describes 4% of variance in disease risk. I think this should be described for all of the reports, to give a sense of the predictive power (along with other statistics).
I also noticed most of the variants were not present in ClinVar (when I was using dbSNP to check the hg19 genome coordinates and reference allele).
As you can see from this link, the percentile above was calculated using 56 SNPs.
I have blood test results uploaded on my PatientsLikeMe profile, and I thought that I had a normal CRP result. However, it appears that I might not have remembered that correctly, and I might need to wait until my next checkup to see if I can test my CRP level. However, this is something where I think it would be relatively easy to show if being at the 99% percentile substantially affects your observed CRP levels (or whether there are limits to what this score represents).
As you can see from this link, the percentile above was calculated using 6 SNPs. Nebula predicted that I had a homozygous variant for 1 SNP and heterozygous variant for 1 SNP (and reference genotypes for the other 4 variants). However, all of these variants looked OK in my higher coverage Veritas WGS data.
I believe that I have been previously reported to be at higher risk for restless leg syndrome, but that might have actually been for deep vein thrombosis / venous thromboembolism in the earlier 23andMe reports (before the FDA required approval for a more select set of results). I do sometimes have difficult sitting perfectly still at night. However, this does't happen all of the time, and I have never been diagnosed by a doctor for having this condition. So, I would currently lean towards saying that I don't have restless leg syndrome.
For the 2 sets of SNPs that I checked (for alopecia areata and restless leg syndrome), I also visualized my Genos Exome alignment. However, most of the variants were not covered by sequencing of coding regions.
In general, the journals where these results are published may make some readers think the results are useful. However, being able to publish a result in a prestigious journal doesn't mean the associations are predictive enough to be clinically meaningful. Also, even with a more subtle association, being in a prestigious journal doesn't necessarily mean the result can be reproduced. For example, there are retractions in high impact journals (you can see some in this blog post), and there are objectively wrong conclusions in papers that haven't been retracted (and the science-wide error rate is mentioned in this blog post). I don't want to cause unnecessarily alarm, but I think it is important to emphasize that time and large sample sizes (and independent validation) are needed to become comfortable with using genetic results to guide your medical treatment.
Nebula does provide a warning: "Disclaimer: Nebula Library is for research, information, and educational use only. This information is not medical advice, nor is it intended to be used for any diagnostic purpose. Please seek the assistance of a health care provider with any questions regarding your health." However, I think this is easy to miss and the importance may not be fully understood among all customers.
Additional Note #1: I tried to provide a review for Nebula on Trustpilot, but I have encountered some difficulties. You can see a screenshot of the current review (that was not accepted) here. While I still haven't gotten a response for the last attempt to submit a review and get an explanation of what I need to change. While I am not certain if this is the cause, the link that I received to submit a review initially created a review under another name. To be fair, Nebula did pay me the $10 Amazon gift card (even when I provided a screenshot of a 2-star review, when most are 4- or 5-star reviews), and I hope that this review can eventually be posted (in which case, I will provide a link to that review, instead of this longer explanation).
Additional Note #2: You can see my report to FDA MedWatch (MW5093887) in MAUDE here. I received an acknowledgement via mail for another report, but I just looked for this report after waiting a while.
Update Log:
8/4/2019 - public post date
8/5/2019 - add update about Nebula research library
8/6/2019 - minor changes
8/10/2019 - add link for 23andMe SNP chip versus WGS concordance
8/14/2019 - minor changes
8/15/2019 - minor changes; add box around APOE variant
8/16/2019 - minor changes
8/17/2019 - revise title (to better emphasize importance, but also presentation of just my own data)
8/16/2019 - minor changes
11/26/2019 - add arrows to Nebula samples in IGV screenshots
1/26/2020 - add screenshot of Trustpilot review
2/24/2020 - mention that Nebula is currently offering higher coverage sequencing
3/19/2020 - add concerns about Nebula Library (to match what I described in my MedWatch report)
3/20/2020 - add links to SNP details for selected library results
3/22/2020 - add link to other PRS post
4/18/2020 - add notes about checking RA post
4/24/2020 - add notes about FDA MedWatch submission
4/26/2020 - minor changes
7/6/2020 - minor changes "Polygenic Risk Score" label, for the last part of the blog post.
8/5/2019 - add update about Nebula research library
8/6/2019 - minor changes
8/10/2019 - add link for 23andMe SNP chip versus WGS concordance
8/14/2019 - minor changes
8/15/2019 - minor changes; add box around APOE variant
8/16/2019 - minor changes
8/17/2019 - revise title (to better emphasize importance, but also presentation of just my own data)
8/16/2019 - minor changes
11/26/2019 - add arrows to Nebula samples in IGV screenshots
1/26/2020 - add screenshot of Trustpilot review
2/24/2020 - mention that Nebula is currently offering higher coverage sequencing
3/19/2020 - add concerns about Nebula Library (to match what I described in my MedWatch report)
3/20/2020 - add links to SNP details for selected library results
3/22/2020 - add link to other PRS post
4/18/2020 - add notes about checking RA post
4/24/2020 - add notes about FDA MedWatch submission
4/26/2020 - minor changes
7/6/2020 - minor changes "Polygenic Risk Score" label, for the last part of the blog post.












This comment has been removed by a blog administrator.
ReplyDelete