I haven't had a whole lot of personal experience with Polygenic Risk Score (PRS) estimates, so I thought it was interesting when I found a couple options for re-analysis of my own genomics data (for selected examples):
| Association | SNP chip (impute.me) (Folkersen et al. 2020) |
Other Re-Analysis Options |
23andMe Results |
|---|---|---|---|
| Type 2 Diabetes (No) |
(Type 2 Diabetes, 146 variants) Average / Above Average (23andMe-V3, 12/19) Average / Above Average (AncestryDNA, 12/19) |
MySeq 1.000 risk ratio [error] (Nebula lcWGS) 0.955 risk ratio (Genos Exome, 3 variants) 1.089 risk ratio (Veritas WGS, 6 variants) |
"Typical Risk" of 23% (directly from 23andMe, PRS with 1,244 loci) [actually, slightly lower than normal] Reduces to less than 1% when age, height, weight, fast food consumption, and exercise rate are taken into consideration (also from 23andMe) |
| Ulcerative Colitis (once, so I think really "no") |
(23 variants, and 116 variants) Both Below Average and Above Average Risk, for different PRS (23andMe-V3, 12/19) Both Below Average and Above Average Risk, for different PRS (AncestryDNA, 12/19) |
||
| Anxiety Disorder (Yes, but getting better) |
(6 variants) Average / Above Average (23andMe-V3, 12/19) Average / Above Average (AncestryDNA, 12/19) |
||
| Migraine (Periodic) |
(26 variants, and 21 variants) 2 PRS (Average and Above Average) (23andMe-V3, 12/19) 2 PRS (Average and Above Average) (AncestryDNA, 12/19) |
||
| Eye Color (Light Brown) |
DNA.land Likely to have Brown Eyes (23andMe-V3, 12/19) Likely to have Brown Eyes (23andMe-V3_V5, 12/19) Likely to have Brown Eyes (AncestryDNA, 12/19) |
23andMe reports that I am expected to have "brown or hazel eyes" based upon 1 SNP (rs12913832) | |
| Hair Color (Light Brown) |
See Below (Roughly 25% Red and 50% Blonde) |
For "Light or Dark Hair", 23andMe reports that I have "Likely Dark" Hair (using 42 SNPs) For "Red Hair" 23andMe reports that I am "Unlikely to have red hair" (using 3 MC1R SNPs: rs1805007, rs1805008, and another custom MCR1 probe) |
|
| Height (180 cm) |
See Below | DNA.land 171 cm: "Likely Taller than Average" (23andMe-V3, 12/19) 171 cm: "Likely Taller than Average" (23andMe-V3_V5, 12/19) 171 cm: "Likely Taller than Average" (AncestryDNA, 12/19) |
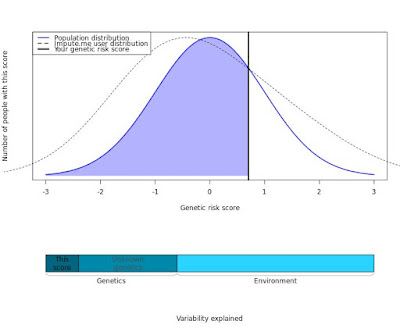
Individual SNP risks were reported (from impute.me). While I had a bit of a hard time finding the precise overall risk estimate (without trying to sum / multiply separate risks), this might be OK in terms of getting a sense of whether I was an outlier or not. For example, being above or below average for "Type 2 Diabetes" seemed to vary (unless you say most people were under something like a null distribution for "average" risk). In other words, I thought the following plots (which you could see for various traits) were interesting:
impute.me Type 2 Diabetes PRS (23andMe V3)
impute.me Ulcerative Colitis (1st entry, 23andMe V3)
impute.me Ulcerative Colitis (2nd entry, 23andMe V3)
impute.me Anxiety Disorder PRS (23andMe V3)
impute.me Migraine-Broad PRS (23andMe V3)
impute.me Migraine PRS (23andMe V3)
I thought the anxiety disorder result was interesting for 2 reasons. First, I have had issues with anxiety problems (for example, you can click here for notes, even though they are primarily related to PatientsLikeMe). Second, notice the environmental component is larger than the genetics component. This matches my concerns that I expressed in this review of "blueprint". For example, I would say the predictive power from birth has some notable limitations (such as difficulties in the need to take medication at any given point in your life).
While I am not sure if the exact right term was used (since I thought "Ulcerative Colitis" was a condition, rather than a symptom). However, I was hospitalized for Ulcerative Colitis (even though that was a one time occurrence caused from E. coli with Shiga toxin).
I also get migraines.
I don't have Type 2 Diabetes, but I provided that because I also had other PRS results to compare. Similarly, if others have suggestions where I can quickly compare to the impute.me PRS results, please let me know and I would be very happy to add them!
For example, I did add DNA.land (and 23andMe) Eye Color and Height based upon a Twitter response. While I think height is one of the more heritable traits, DNA.land couldn't guess my actual height within a few inches (and there is a noticeable spread of points for the impute.me plot above). Even though DNA.land gave lower confidence to other predictions, I would say these have been "fair" rather than "high" confidence (and everything else probably should have been "low" confidence). I am close to the diagonal for the impute.me plot, but I don't know if the scale is 1:1. For example, my DNA.land height prediction was off by 3-4 inches. However, to be fair, note that the highest and lowest percentiles for high don't have overlap (there are not any points in the upper-left or bottom-right regions of the scatter plot, even though those make up a smaller fraction of the population).
For comparison, here is the distribution of score for DNA.land (where my true height was greater than anything on the density distribution - perhaps because this was height scaled for female percentiles?):
For impute.me, the predicted hair color shows blondness on the x-axis and redness on the y-axis. The cyan circle is my actual color (which I filled in), and the while circle is my predicted color. I think my hair color used to be lighter than it is now (and I think the shade that I reported for myself was a bit too dark), so that is closer to the genetic prediction (perhaps half-way between).
It may be worth noting that 23andMe could predict that I had brown hair and eyes (although I think that covers most people and you need the more rare traits to better calculate accuracy - for example, Francis Collins said that his 23andMe report indicated he had brown eyes when he really had blue eyes, at least 10 years ago).
Again, for comparison, here is the distribution of DNA.land scores for eye color:
I didn't add the AncestryDNA density plots since they looked qualitatively similar to the 23andMe V3 plots (and, on another computer, I had an issue with the percent variance explained appearing in a pie chart that was harder to read). I also originally intended to test my updated 23andMe genotypes (V3+V5), but I got an error saying that data was already uploaded (from my V3 chip). However, perhaps I can test those results later, and see if they are still similar.
With a $5 donation, the turn-around time for processing was 1-3 days.
For Genos Exome and Veritas WGS data, I used the BWA-MEM Re-Aligned GATK Variant calls. However, I think the main conclusion from looking at my diabetes results was that I was of average risk, and I don't believe my own genetic diabetes PRS risk assessment was great without taking additional factors into consideration (for 23andMe, that was a difference between 23% and 1%, after considering BMI, diet, and exercise).
This essentially matches Supplementary Figure S12 for this paper (whose title I respectfully believe can give the reader the wrong impression, and there is at least one objective error that I believe needs to be corrected), where absolute risk explained was usually very low (usually explaining less than 15% of the variation for a trait). You can also see that the variability explained by "this score" for the impute.me PRS above is estimated to be less than half of the genetic component.
I think the preprint by Brockman et al. 2021 might also have some additional relevant information for this discussion.
In somewhat different contexts, you can also see some notes / concerns about percentiles / indices in the posts on Nebula and basepaws lcWGS results.
Change Log:
12/5/2019 - public post
12/7/2019 - add DNA.land results based upon Twitter reply from Debbie Kennett; revise wording in post
12/8/2019 - mention possible scaling for female height; also fix date for previous log entry.
6/25/2020 - add links to posts with Nebula and basepaws results. Minor formatting changes.
7/7/2020 - add reference to impute.me paper
4/22/2021 - add reference to another paper
2/4/2024 - change column labels to be more precise















This comment has been removed by a blog administrator.
ReplyDelete