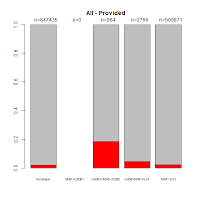
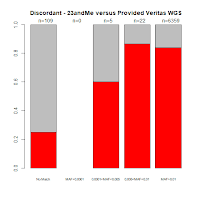
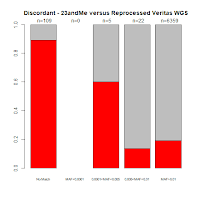
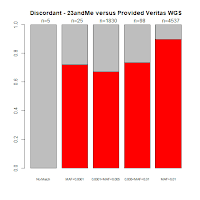
Comments with Successful Follow-Up:
Kreutz et al. 2020 (Bioinformatics) - correct typo in title after PubPeer comment
Martin et al. 2019 (Nature Genetics) - formal erratum, and earlier PubPeer comment (one typo, one suggestion that Figure S12 makes absolute accuracy more clear)
Jonsson et al. 2019 (Nature) - formal correction following article comment
Weedon et al. 2019 (bioRxiv pre-print) - extra data (including my own) was added after comment
Zhang et al. 2019 (PLOS Computational Biology) - I had a comment that I believe was able to be corrected before the final version of the paper (although the paper did later have a formal correction)
Pique-Regi et al. 2019 (bioRxiv pre-print) - discussion helped fix links in pre-print
BLAST reference issue reported to NCBI (caused from a number of different papers no recognizing cross contamination, I believe e-mail was sent showing incorrect annotations for a limited number of example sequences). It looks like problem was not permanently fixed (due to similar problems with new data from other studies). However, through whatever mechanism, top PhiX BLAST hits that were incorrect were removed - not sure if it was the actual cause for correction, but I posted this on Biostars when it was a problem
Multiple but Primarily Minor Errors (not currently fixed):
Weedon et al. 2019 (bioRxiv pre-print) - extra data (including my own) was added after comment
Zhang et al. 2019 (PLOS Computational Biology) - I had a comment that I believe was able to be corrected before the final version of the paper (although the paper did later have a formal correction)
Pique-Regi et al. 2019 (bioRxiv pre-print) - discussion helped fix links in pre-print
BLAST reference issue reported to NCBI (caused from a number of different papers no recognizing cross contamination, I believe e-mail was sent showing incorrect annotations for a limited number of example sequences). It looks like problem was not permanently fixed (due to similar problems with new data from other studies). However, through whatever mechanism, top PhiX BLAST hits that were incorrect were removed - not sure if it was the actual cause for correction, but I posted this on Biostars when it was a problem
Multiple but Primarily Minor Errors (not currently fixed):
Yizhak et al. 2019 - blog post (errors and suggestions) and PubPeer comment (at least one clear typo); I also posted an eLetter describing the 2 most clear errors; Science paper
Arguably Gives Reader Wrong Impression:
Li et al. 2021 - there are a number of things that I am understanding better by participating in the discussion process. Also, my original intention was to add a Disqus-style comment on the journal article, but that was not provided. So, that is why I posted a PubPeer comment.
Essentially, I think I would prefer a SNP chip (or Exome, higher coverage WGS, Amplicon-Seq, etc.) over lcWGS (especially the 0.5x to 1x presented in this paper), and I think the value of directly measured genotypes for SNP chips is being underestimated. I agree with lcWGS imputations often being preferable to SNP chip imputations, but I think inclusion of imputed SNP chip genotypes may not be made sufficiently clear to the broader audience.
I also mention problems with consumer lcWGS products.
Maya et al. 2020 - if I understand the paper correctly, the study does not directly investigate COVID-19 infections (they are looking for associations near ACE2 or TMPRSS2, but in other contexts). Please scroll to the bottom to see my comment.
Nature summary of PLOS ONE paper - image shows input (not output) of encoding and attempted recovery; unlike Nature research reports, I didn't see a Disqus comment system (but I did mention something on Twitter)
Homburger et al. 2019 - I had enough problems with my lcWGS data (which was exceptionally low coverage for my Color data) that I would not recommend it's use. I realize that sample size is limited (just my own data). However, I posted my concerns on a pre-print for this article. I think this is borderline for the previous category ("Data Contradictory to Main Conclusions", more appropriate for a retraction), but I would need more data to conclude that. I also posted a similar response on PubPeer.
I think this blog post on being able to self-identify myself can give some sense of the concerns about the very low coverage WGS reads that I received from Color.
Minor Typos:
Nobles et al. 2019 - PubPeer comment
Gambardella and di Bernardo 2019 - PubPeer comment for typo
Guo et al. 2019 - comment about typo in PLOS comment system.
Chen et al. 2019 - PubPeer comment
Qin et al. 2018 - Disqus journal comment
Maya et al. 2020 - if I understand the paper correctly, the study does not directly investigate COVID-19 infections (they are looking for associations near ACE2 or TMPRSS2, but in other contexts). Please scroll to the bottom to see my comment.
Nature summary of PLOS ONE paper - image shows input (not output) of encoding and attempted recovery; unlike Nature research reports, I didn't see a Disqus comment system (but I did mention something on Twitter)
Homburger et al. 2019 - I had enough problems with my lcWGS data (which was exceptionally low coverage for my Color data) that I would not recommend it's use. I realize that sample size is limited (just my own data). However, I posted my concerns on a pre-print for this article. I think this is borderline for the previous category ("Data Contradictory to Main Conclusions", more appropriate for a retraction), but I would need more data to conclude that. I also posted a similar response on PubPeer.
I think this blog post on being able to self-identify myself can give some sense of the concerns about the very low coverage WGS reads that I received from Color.
Singer et al. 2019 - eDNA paper with multiple comments indicating concerns, and I specifically found there were extra PhiX reads in the NovaSeq samples. As mentioned in the comment, I have an "answer" on a Biostars discussion more broadly related to PhiX (with a temporary success story about the BLAST database mentioned later in this post).
The author provided a helpful response indicating that the PhiX reads should be removed for DADA2 for downstream analysis. However, I think we both agree that PhiX spike-ins don't have a barcode. So, if you view that as extra cross-contamination in some samples, then this leaves the question of what else could be in the NovaSeq samples that might be harder to flag as something that needs to be removed. Time permitting, I am still looking into this.
Minor Typos:
Yang et al. 2021 - PubPeer comment
Giansanti et al. 2020 - F1000 Research comment
Kreutz et al 2020 - PubPeer comment (typo in title)
Giansanti et al. 2020 - F1000 Research comment
Kreutz et al 2020 - PubPeer comment (typo in title)
Yuan and Bar-Joseph 2019 - PubPeer comment about MSigDB citation (as GSEA)
Nobles et al. 2019 - PubPeer comment
Gambardella and di Bernardo 2019 - PubPeer comment for typo
Guo et al. 2019 - comment about typo in PLOS comment system.
Chen et al. 2019 - PubPeer comment
Qin et al. 2018 - Disqus journal comment
Chen et al. 2013 - comment about typos in PLOS comment system
Doorslaer and Burk 2010 - PubPeer comment
Other:
Börjesson et al. 2022 - PubPeer question/comment
Andrews et al. 2021 - PubPeer question (because Nature Methods doesn't have a Disqus comment system; also, public acknowledgement of misunderstanding on my part)
Antipov et al. 2020 - PubPeer comment (comments left in Word document for Supplemental Figure S2)
Robitaille et al. 2020 - PubPeer comment (question, posted there due to lack of comment system)
Older et al. 2019 - comment in PLOS comment system.
Young 2019 - comment in PLOS comment system
Choonoo et al. 2019 - comment in PLOS comment system
Paz-Zulueta et al. 2018 - PubPeer comment
Mirabello et al. 2017 - PubPeer comment
Shen-Gunther et al. 2017 - PubPeer comment
Robinson and Oshlack 2010 - PubPeer question (in the interests of fairness, I believe this reflects misunderstanding on my part)
Munoz et al. 2003 - PubPeer question (I didn't find any errors in the paper, but I wondered why data was presented in a particular way, and journal didn't have a comment system)
I also have some notes on limits to precision in genomics results available to the public (which I would probably define more as a "product" than a "paper), among this collection of posts.
Given it is harder for me to find these (compared to my Disqus comments), I will probably add more in the future (but the comments in the "other" section are not necessarily bad - I will just probably forget them if I don't save a link here). For example, I believe increased participation in such comment systems may make them as effective as a "minor comment" (particularly if the journal doesn't go back and change the original publication after a formal correction).
Checking / Correcting Citations of My 1st Author Papers
Silva et al. 2022 - journal comment
Xu et al. 2021 - PubPeer comment
Wojewodzic and Lavender et al. 2021 - preprint comment (hopefully, can be corrected before peer-reviewed publication)
Roudko et al. 2020 - journal comment (please scroll down, past references, to see comment)
Nayak et al. 2019 - Disqus comment (on pre-print)
Borchmann et al. 2019 - Disqus comment (on pre-print)
Youness and Gad 2019 - PubPeer comment
Muller et al. 2019 - Twitter comment
Bogema et al. 2018 - PubPeer comment
Einarsdottir et al. 2018 - PubPeer comment
Pranckeniene et al. 2018 - journal comment (please scroll down, past references, to see comment)
Debniak et al. 2018 - journal comment
Hu et al. 2016 - PubPeer comment (probably due to confusion on Bioconductor page)
Wong and Chang 2015 - PubPeer comment
Wockner et al. 2015 - PubPeer comment (more of a true comment, than post-publication review for error)
As mentioned in a couple other posts (general and COH-specific), I am trying to correct previous errors in my papers, and I think having 4 first-author (or equivalent) papers in 2013 was probably not ideal (in terms of needing to take more time to carefully review each paper).
However, I also want to show a relatively long list of corrections / retractions (even though they still make up a subset of the total public records), with some greater emphasis on high impact papers and/or papers with a large number of authors. To be fair, I may not know the details of the individual corrections or retractions. However, I hope this will cause future researchers to be brave and responsible and report / correct problems as soon as they discover them. As a best case scenario, I believe that it is important for prestigious labs to lead by example (so that everybody represents themselves fairly and finds the best long-term fit). We do not want to encourage people to hesitate reporting problems out of fear that they will lose funding and/or collaborations with peers (and/or those that do worse work but less frequently admit errors and/or oversell results will be chosen over those who present more realistic expectations). I am very confident these are achievable goals, but I think some topics may need relatively more frequent discussions.
Other Positive Examples of Corrections:
I am trying to collect examples for high-impact papers/journals and/or consortiums.
Correction for Wang et al. 2020 (Nature Communications paper, The Genome Aggregation Database Consortium paper)
Doorslaer and Burk 2010 - PubPeer comment
Other:
Börjesson et al. 2022 - PubPeer question/comment
Andrews et al. 2021 - PubPeer question (because Nature Methods doesn't have a Disqus comment system; also, public acknowledgement of misunderstanding on my part)
Antipov et al. 2020 - PubPeer comment (comments left in Word document for Supplemental Figure S2)
Robitaille et al. 2020 - PubPeer comment (question, posted there due to lack of comment system)
Older et al. 2019 - comment in PLOS comment system.
Young 2019 - comment in PLOS comment system
Choonoo et al. 2019 - comment in PLOS comment system
Paz-Zulueta et al. 2018 - PubPeer comment
Mirabello et al. 2017 - PubPeer comment
Shen-Gunther et al. 2017 - PubPeer comment
Robinson and Oshlack 2010 - PubPeer question (in the interests of fairness, I believe this reflects misunderstanding on my part)
Munoz et al. 2003 - PubPeer question (I didn't find any errors in the paper, but I wondered why data was presented in a particular way, and journal didn't have a comment system)
I also have some notes on limits to precision in genomics results available to the public (which I would probably define more as a "product" than a "paper), among this collection of posts.
Given it is harder for me to find these (compared to my Disqus comments), I will probably add more in the future (but the comments in the "other" section are not necessarily bad - I will just probably forget them if I don't save a link here). For example, I believe increased participation in such comment systems may make them as effective as a "minor comment" (particularly if the journal doesn't go back and change the original publication after a formal correction).
Checking / Correcting Citations of My 1st Author Papers
Silva et al. 2022 - journal comment
Xu et al. 2021 - PubPeer comment
Wojewodzic and Lavender et al. 2021 - preprint comment (hopefully, can be corrected before peer-reviewed publication)
Roudko et al. 2020 - journal comment (please scroll down, past references, to see comment)
Nayak et al. 2019 - Disqus comment (on pre-print)
Borchmann et al. 2019 - Disqus comment (on pre-print)
Youness and Gad 2019 - PubPeer comment
Muller et al. 2019 - Twitter comment
Bogema et al. 2018 - PubPeer comment
Einarsdottir et al. 2018 - PubPeer comment
Pranckeniene et al. 2018 - journal comment (please scroll down, past references, to see comment)
Debniak et al. 2018 - journal comment
Hu et al. 2016 - PubPeer comment (probably due to confusion on Bioconductor page)
Wong and Chang 2015 - PubPeer comment
Wockner et al. 2015 - PubPeer comment (more of a true comment, than post-publication review for error)
As mentioned in a couple other posts (general and COH-specific), I am trying to correct previous errors in my papers, and I think having 4 first-author (or equivalent) papers in 2013 was probably not ideal (in terms of needing to take more time to carefully review each paper).
However, I also want to show a relatively long list of corrections / retractions (even though they still make up a subset of the total public records), with some greater emphasis on high impact papers and/or papers with a large number of authors. To be fair, I may not know the details of the individual corrections or retractions. However, I hope this will cause future researchers to be brave and responsible and report / correct problems as soon as they discover them. As a best case scenario, I believe that it is important for prestigious labs to lead by example (so that everybody represents themselves fairly and finds the best long-term fit). We do not want to encourage people to hesitate reporting problems out of fear that they will lose funding and/or collaborations with peers (and/or those that do worse work but less frequently admit errors and/or oversell results will be chosen over those who present more realistic expectations). I am very confident these are achievable goals, but I think some topics may need relatively more frequent discussions.
Other Positive Examples of Corrections:
I am trying to collect examples for high-impact papers/journals and/or consortiums.
Correction for Collins et al. 2020 (Nature paper, The Genome Aggregation Database Consortium paper)
Correction for Karczewski et al. 2020 (Nature paper, The Genome Aggregation Database Consortium paper)
Correction for Minikel et al. 2020 (Nature paper, The Genome Aggregation Database Consortium paper)
Correction for Wang et al. 2020 (Nature Communications paper, The Genome Aggregation Database Consortium paper)
Correction for Whiffin et al. 2020 (Nature Communications paper, The Genome Aggregation Database Consortium paper)
Correction for Cortés-Ciriano et al. 2020 (Nature Genetics paper)
Retraction for Cho et al. 2019 (Science / Nobel Laurete paper: I learned about from Retraction Watch)
Retraction for Wei and Nielson 2019 (Nature Medicine paper)
--> Post-publication view indicated on Twitter in advance
--> Also includes retraction of "News & Views" article
Correction to Kelleher et al. 2019 (Nature Genetics paper)
Correction to Grishin et al. 2019 (Nature Biotechnology paper)
Publisher Correction to Exposito-Alonso et al. 2019 (Nature paper, as well as "500 Genomes Field Experiment Team" consortium paper)
Correction to Bolyen et al. 2019 - (Nature Biotechnology paper; correction for QIIME II paper)
Correction to Kruche et al. 2019 - (Nature Biotechnology paper; GA4GH Small Variant Benchmarking)
Retraction to Kaidi et al. 2019 (Nature paper; positive in the sense author resigned and admitted fabrication, rather than denying fabrication; I learned about from Retraction Watch)
Retraction to Kaidi et al. 2019 (Science paper; positive in the sense author resigned and admitted fabrication, rather than denying fabrication; I learned about from Retraction Watch)
Correction to Ravichandran et al. 2019 (Genetics in Medicine paper; added conflicts of interest)
Erratum to Jiang et al. 2019 (one affiliation issue with Nature Communications paper with a lot of authors)
Correction to Ferdowsi et al. 2018 - (mixed up Figures; Scleroderma Clinical Trials Consortium Damage Index Working Group paper)
Correction to Sanders et al. 2018 - (Nature Neuroscience paper; Whole Genome Sequencing for Psychiatric Disorders (WGSPD) consortium
2 corrections to Gandolfi et al. 2018 paper on cat SNP chip array
Corrigendum to Oh et al. 2018 (99 Lives Consortium paper)
Correction to (a different) Gandolfi et al. 2018
Correction to Vijayakrishnan 2018 (PRACTICAL Consortium included on authors list)
Correction to Matejcic et al. 2018 (another PRACTICAL Consortium paper)
Correction to Went et al. 2018 (another PRACTICAL Consortium paper)
Correction to Schumacher et al. 2018 (Nature Genetics paper, several consortium listed as authors)
Correction to Mancuso et al. 2018 (another PRACTICAL Consortium paper)
Correction to Armenia et al. 2018 (affiliation correction, Nature genetics paper, Stand Up To Cancer, SU2C, Consortium paper)
Withdraw of Werling 2018 Review (I learned about from Retraction Watch)
Correction to Sud et al. 2017 (another PRACTICAL Consortium paper)
Retraction and Replacement of Favini et al. 2017 (JAMA paper ; I learned about from Retraction Watch)
Corrigendum to McHenry et al. 2017 (Nature Neuroscience paper; I learned about from Retraction Watch)
2 Erratums to Teng et al. 2016 (Genome Biology paper; I found from this Tweet, describing error identified and corrected by another scientist from post-publication review; one Erratum is really a subset of the other)
Correction to Nik-Zainal et al. 2016 (Nature paper; typo not fixed in on-line article)
Erratum to Aberdein et al. 2016 (another 99 Lives Consortium paper)
2 corrections to Vinik 2016 (NEJM paper; I learned about from Retraction Watch)
Retraction of Jia et al. 2016 (Nature Chemistry paper + Nobel Laurate Author; I learned about from Retraction Watch)
Retraction of Colla et al. 2016 (JAMA paper; I learned about from Retraction Watch)
Erratum to Kocher et al. 2015 (Genome Biology paper)
Retraction of Zhang et al. 2015 (Nature paper; I learned about from Retraction Watch)
2 Retractions (?) to Akakin et al. 2015 (Journal of Neurosurgery paper; I learned about from Retraction Watch)
Addendum to Koh et al. 2015 (Nature Methods paper; figures were reversed, creating mirror images)
Retraction and Replacement for Hollon et al. 2014 (JAMA Psychiatry paper; I learned about from Retraction Watch)
Retraction to Kitambi et al. 2014 (Cell paper; I learned about from Retraction Watch)
Correction to Huang et al. 2014 (Nature paper; I learned about from Retraction Watch)
Retraction and Replacement of Li et al. 2014 (The Lancet; I learned about from Retraction Watch)
Retraction and Replacement of Siempos et al. 2014 (The Lancet Respiratory Medicine paper; I learned about from Retraction Watch)
Correction to Xu et al. 2014 (eLife paper; I learned about from Retraction Watch)
Retraction of Lortez et al. 2014 (Science paper; I learned about from Retraction Watch)
Retraction of De la Herrán-Arita et al. 2014 (Science Translational Medicine paper; I learned about from Retraction Watch)
Retraction for Dixson et al. 2014 (PNAS paper; I learned about from Retraction Watch)
Retraction of Garcia-Serrano and Frankignoul 2014 (Nature Geoscience paper; I learned about from Retraction Watch)
Retraction of Amara et al. 2013 (JEM paper; I learned about from Retraction Watch)
Retraction of Maisonneuve et al 2013 (Cell paper; I learned about from Retraction Watch)
Retraction of Yi et al. 2013 (Cell paper; I learned about from Retraction Watch)
Retraction of Nandakumar et al. 2013 (PNAS paper; I learned about from Retraction Watch)
Retraction of Venters and Pugh 2013 (Nature paper; I learned about from Retraction Watch, describing 6th Nature retraction in 2014)
Retraction of Maisonneuve et al 2011 (PNAS paper; I learned about from Retraction Watch)
Retraction of Frede et al. 2011 (Blood paper; I learned about from Retraction Watch)
Retraction to Olszewski et al. 2010 (Nature paper; mentioned in this Retraction Watch article)
Correction to Werren et al. 2010 (Science paper; correction not actually indexed in PubMed?)
Retraction of Bill et al. 2010 (Cell paper; I learned about from Retraction Watch)
Retraction to Wang et al. 2009 (Nature paper; I learned about from Retraction Watch)
Retraction of Litovchick and Szostak 2008 (PNAS paper + Nobel Laurate Author; I learned about from Retraction Watch)
Retraction of Okada et al. 2006 (Science paper; I learned about from Retraction Watch)
Withdraw of Ruel et al. 1999 (17 years post publication; JBC paper; I learned about from Retraction Watch)
I think this last category is really important (even though it is still no where near complete - it is just some representative examples that I know about).
Change Log:
8/26/2019 - public post date
8/27/2019 - add sentence about increased comment participating being more like formal comment (in some situations)
8/29/2019 - minor changes
8/29/2019 - added examples tagged with "doing the right thing" in Retraction Watch. Thank you very much to Dave Fernig!
8/30/2019 - changed tense of previous entry of change log (from future tense to past tense)
9/2/2019 - add another positive example of comments improving pre-print
9/5/2019 - fix typo; minor changes
9/17/2019 - all 2019 correction, which I remembered from this tweet
9/18/2019 - fix typo; add multiple other citations
9/20/2019 - add PLOS Genetics correction as successful example
9/30/2019 - add recent Nature / Nature Medicine corrections
10/8/2019 - add Nature Medicine CCR5 official retraction and Nature Biotechnology / Nature Genetics corrections
10/18/2019 - add another PLOS ONE comment, PhiX example, and started list of incorrect citations for my 1st author papers
10/24/2019 - add PubPeer comment and eDNA paper
10/25/2019 - add PubPeer comment
10/28/2019 - add more PubPeer / citation comments
10/30/2019 - add Twitter comment
11/8/2019 - add a couple more corrections
11/10/2019 - revise sentences about being "brave"
11/11/2019 - add another PubPeer comment
12/2/2019 - add lcWGS concern + PLOS "filleted" typo
12/10/2019 - add positive example for formal Nature correction
12/24/2019 - add another PubPeer comment
12/30/2019 - add another PubPeer comment
1/2/2020 - add PubPeer question + Science retraction
1/14/2019 - add another PubPeer comment
1/29/2020 - add another Disqus comment
4/24/2020 - add comment for citation of one of my papers
5/15/2020 - minor changes
6/4/2020 - add PubPeer entry for article with typo in title
6/9/2020 - add COVID-19 paper that I believe uses confusing wording
6/15/2020 - add F1000 Research comment
6/25/2020 - minor formatting changes
Correction for Cortés-Ciriano et al. 2020 (Nature Genetics paper)
Retraction for Cho et al. 2019 (Science / Nobel Laurete paper: I learned about from Retraction Watch)
Retraction for Wei and Nielson 2019 (Nature Medicine paper)
--> Post-publication view indicated on Twitter in advance
--> Also includes retraction of "News & Views" article
Correction to Kelleher et al. 2019 (Nature Genetics paper)
Correction to Grishin et al. 2019 (Nature Biotechnology paper)
Publisher Correction to Exposito-Alonso et al. 2019 (Nature paper, as well as "500 Genomes Field Experiment Team" consortium paper)
Correction to Bolyen et al. 2019 - (Nature Biotechnology paper; correction for QIIME II paper)
Correction to Kruche et al. 2019 - (Nature Biotechnology paper; GA4GH Small Variant Benchmarking)
Retraction to Kaidi et al. 2019 (Nature paper; positive in the sense author resigned and admitted fabrication, rather than denying fabrication; I learned about from Retraction Watch)
Retraction to Kaidi et al. 2019 (Science paper; positive in the sense author resigned and admitted fabrication, rather than denying fabrication; I learned about from Retraction Watch)
Correction to Ravichandran et al. 2019 (Genetics in Medicine paper; added conflicts of interest)
Erratum to Jiang et al. 2019 (one affiliation issue with Nature Communications paper with a lot of authors)
Correction to Ferdowsi et al. 2018 - (mixed up Figures; Scleroderma Clinical Trials Consortium Damage Index Working Group paper)
Correction to Sanders et al. 2018 - (Nature Neuroscience paper; Whole Genome Sequencing for Psychiatric Disorders (WGSPD) consortium
2 corrections to Gandolfi et al. 2018 paper on cat SNP chip array
Corrigendum to Oh et al. 2018 (99 Lives Consortium paper)
Correction to (a different) Gandolfi et al. 2018
Correction to Vijayakrishnan 2018 (PRACTICAL Consortium included on authors list)
Correction to Matejcic et al. 2018 (another PRACTICAL Consortium paper)
Correction to Went et al. 2018 (another PRACTICAL Consortium paper)
Correction to Schumacher et al. 2018 (Nature Genetics paper, several consortium listed as authors)
Correction to Mancuso et al. 2018 (another PRACTICAL Consortium paper)
Correction to Armenia et al. 2018 (affiliation correction, Nature genetics paper, Stand Up To Cancer, SU2C, Consortium paper)
Withdraw of Werling 2018 Review (I learned about from Retraction Watch)
Correction to Sud et al. 2017 (another PRACTICAL Consortium paper)
Retraction and Replacement of Favini et al. 2017 (JAMA paper ; I learned about from Retraction Watch)
Corrigendum to McHenry et al. 2017 (Nature Neuroscience paper; I learned about from Retraction Watch)
2 Erratums to Teng et al. 2016 (Genome Biology paper; I found from this Tweet, describing error identified and corrected by another scientist from post-publication review; one Erratum is really a subset of the other)
Correction to Nik-Zainal et al. 2016 (Nature paper; typo not fixed in on-line article)
Erratum to Aberdein et al. 2016 (another 99 Lives Consortium paper)
2 corrections to Vinik 2016 (NEJM paper; I learned about from Retraction Watch)
Retraction of Jia et al. 2016 (Nature Chemistry paper + Nobel Laurate Author; I learned about from Retraction Watch)
Retraction of Colla et al. 2016 (JAMA paper; I learned about from Retraction Watch)
Erratum to Kocher et al. 2015 (Genome Biology paper)
Retraction of Zhang et al. 2015 (Nature paper; I learned about from Retraction Watch)
2 Retractions (?) to Akakin et al. 2015 (Journal of Neurosurgery paper; I learned about from Retraction Watch)
Addendum to Koh et al. 2015 (Nature Methods paper; figures were reversed, creating mirror images)
Retraction and Replacement for Hollon et al. 2014 (JAMA Psychiatry paper; I learned about from Retraction Watch)
Retraction to Kitambi et al. 2014 (Cell paper; I learned about from Retraction Watch)
Correction to Huang et al. 2014 (Nature paper; I learned about from Retraction Watch)
Retraction and Replacement of Li et al. 2014 (The Lancet; I learned about from Retraction Watch)
Retraction and Replacement of Siempos et al. 2014 (The Lancet Respiratory Medicine paper; I learned about from Retraction Watch)
Correction to Xu et al. 2014 (eLife paper; I learned about from Retraction Watch)
Retraction of Lortez et al. 2014 (Science paper; I learned about from Retraction Watch)
Retraction of De la Herrán-Arita et al. 2014 (Science Translational Medicine paper; I learned about from Retraction Watch)
Retraction for Dixson et al. 2014 (PNAS paper; I learned about from Retraction Watch)
Retraction of Garcia-Serrano and Frankignoul 2014 (Nature Geoscience paper; I learned about from Retraction Watch)
Retraction of Amara et al. 2013 (JEM paper; I learned about from Retraction Watch)
Retraction of Maisonneuve et al 2013 (Cell paper; I learned about from Retraction Watch)
Retraction of Yi et al. 2013 (Cell paper; I learned about from Retraction Watch)
Retraction of Nandakumar et al. 2013 (PNAS paper; I learned about from Retraction Watch)
Retraction of Venters and Pugh 2013 (Nature paper; I learned about from Retraction Watch, describing 6th Nature retraction in 2014)
Retraction of Maisonneuve et al 2011 (PNAS paper; I learned about from Retraction Watch)
Retraction of Frede et al. 2011 (Blood paper; I learned about from Retraction Watch)
Retraction to Olszewski et al. 2010 (Nature paper; mentioned in this Retraction Watch article)
Correction to Werren et al. 2010 (Science paper; correction not actually indexed in PubMed?)
Retraction of Bill et al. 2010 (Cell paper; I learned about from Retraction Watch)
Retraction to Wang et al. 2009 (Nature paper; I learned about from Retraction Watch)
Retraction of Litovchick and Szostak 2008 (PNAS paper + Nobel Laurate Author; I learned about from Retraction Watch)
Retraction of Okada et al. 2006 (Science paper; I learned about from Retraction Watch)
Withdraw of Ruel et al. 1999 (17 years post publication; JBC paper; I learned about from Retraction Watch)
I think this last category is really important (even though it is still no where near complete - it is just some representative examples that I know about).
Change Log:
8/26/2019 - public post date
8/27/2019 - add sentence about increased comment participating being more like formal comment (in some situations)
8/29/2019 - minor changes
8/29/2019 - added examples tagged with "doing the right thing" in Retraction Watch. Thank you very much to Dave Fernig!
8/30/2019 - changed tense of previous entry of change log (from future tense to past tense)
9/2/2019 - add another positive example of comments improving pre-print
9/5/2019 - fix typo; minor changes
9/17/2019 - all 2019 correction, which I remembered from this tweet
9/18/2019 - fix typo; add multiple other citations
9/20/2019 - add PLOS Genetics correction as successful example
9/30/2019 - add recent Nature / Nature Medicine corrections
10/8/2019 - add Nature Medicine CCR5 official retraction and Nature Biotechnology / Nature Genetics corrections
10/18/2019 - add another PLOS ONE comment, PhiX example, and started list of incorrect citations for my 1st author papers
10/24/2019 - add PubPeer comment and eDNA paper
10/25/2019 - add PubPeer comment
10/28/2019 - add more PubPeer / citation comments
10/30/2019 - add Twitter comment
11/8/2019 - add a couple more corrections
11/10/2019 - revise sentences about being "brave"
11/11/2019 - add another PubPeer comment
12/2/2019 - add lcWGS concern + PLOS "filleted" typo
12/10/2019 - add positive example for formal Nature correction
12/24/2019 - add another PubPeer comment
12/30/2019 - add another PubPeer comment
1/2/2020 - add PubPeer question + Science retraction
1/14/2019 - add another PubPeer comment
1/29/2020 - add another Disqus comment
4/24/2020 - add comment for citation of one of my papers
5/15/2020 - minor changes
6/4/2020 - add PubPeer entry for article with typo in title
6/9/2020 - add COVID-19 paper that I believe uses confusing wording
6/15/2020 - add F1000 Research comment
6/25/2020 - minor formatting changes
7/30/2020 - MetaviralSPAdes comment
7/30/2020 - add TMM question
11/6/2020 - move eDNA category
11/11/2020 - add Bioinformatics typo correction
12/11/2020 - add PLOS ONE comment
2/2/2021 - add a couple more positive correction notes + BMC Medical Genomics figure label issue
2/3/2021 - add additional corrections for consortium papers published in Nature journals on 5/27/2020
3/13/2021 - add MSigDB citation as GSEA
4/8/2021 - minor changes
4/10/2021 - add lcWGS PubPeer comment
4/14/2021 - add another example of successful post-publication review
7/28/2021 - add Nature Methods multiplet PubPeer question
8/6/2021 - add note to acknowledge my misunderstanding for Nature Methods multiplet PubPeer question
10/30/2021 - try to better explain PhiX issue temporarily fixed by NCBI; also, move a subset of examples to my Google Scholar page (since the RNA-MuTect Science paper eLetter indicating a need for corrections was automatically recognized, but I needed to manually fix some things - this made me realize I may be able to use Google Scholar to help emphasize some post-publication review)
3/15/2022 - add MethReg comment
6/2/2022 - additional COHCAP corrigendum citation references
10/4/2022 - TC-hunter PubPeer question/comment