I wanted to first test application to my own samples, which I thought could at least help with troubleshooting errors and providing another independent sample for comparison (not used for training the method being tested).
I also thought it might help to put this in a blog post, since I think this can provide a concrete example of what might help in working towards labs knowingly collaborate with each other (where large portions of work may otherwise be done in different labs without their each other's knowledge).
Strategy #1: Use On-Target High Coverage GATK Variants
Based upon some earlier analysis (related to the Mayo GeneGuide analysis, but not previously posted), there is some additional decrease in IBD/kinship from a perfect self-identification for my Genos Exome sample (even though I think it may still be acceptable for some applications, such as self-identification for QC and privacy purposes):
| Sample 1 | Sample 2 | Kinship |
| Veritas WGS | Genes for Good SNP chip | 0.499907 |
| Veritas WGS | 23andMe SNP chip | 0.499907 |
| Veritas WGS | Mayo GeneGuide Exome+ | 0.499907 |
| Veritas WGS | Genos Exome (BWA-MEM Re-Aligned, GATK Variants) | 0.459216 |
As you can see in the above link, performance was better for the Mayo GeneGuide gVCF. However, I was not provided FASTQ files or BAM files (even when I paid for the Exome+ data to get the gVCF), and Mayo GeneGuide has been discontinued (and I would usually have considered the Genos Exome to be preferable overall). This may be because my Genos Exome target design was for CDS (protein-coding) regions only.
While it is impossible to conduct the 2nd strategy (using off-target reads) with the Mayo GeneGuide Exome+ data, I can test comparing the 2 Exome samples (Genos and Mayo GeneGuide) for ADMIXTURE ancestry analysis:
There is also public code written for a lab for QC Array (SNP chip) application, which is conceptually similar (although my 23andMe and AncestryDNA SNP chip IBD values were higher than my Genos Exome kinship values). This explains the naming of the table with the 1000 Genomes super-population assignments (to create the ADMIXTURE .pop file).
There is also some code for the above ADMXITURE plots here. Those results were roughly similar with all 5 of the samples tested (Veritas WGS, Genes for Good SNP chip, 23andMe SNP chip, Genos Exome, and Mayo GeneGuide Exome+) with 2,272 1000 Genomes reference samples and 9,423 genotypes.
For comparison, if I just compare my 23andMe SNP chip to the 1000 Genomes SNP chips (using an increased count of 92,540 probes), then this is what my ADMIXTURE ancestry looks like:
Strategy #2a: Use Off-Target Reads for STITCH lcWGS Analysis
For comparison, you can see this blog post about using lcWGS for self-identification (where ~1 million reads had similar performance to the Genos CDS Exome Kinship/IBD calculation).
This leads to some improvement in the kinship between my own samples (~0.479 compared to my 23andMe and AncestryDNA SNP chips).
This estimate is similar to the on-target results (~85% European ancestry). However, my AMR percentage was higher (~7% versus <1%), and my EAS percentage was lower (~1% versus 7%). So, I think this was a little more accurate, but I list some important caveats below. However, if you are looking for 1 majority ancestry assignment (and placing less emphasis on <10-20% assignments), then they are essentially the same (and using STITCH considerably increases the run-time).
Importantly, you can also see my lcWGS ancestry analysis here (but for chromosome painting rather than ADMIXTURE, where chromosome painting requires more markers), and you can see my full SNP chip results (mostly for 23andMe) here. So, I was a surprised that the larger number of 23andMe SNPs at the end of the 1st section didn't have an ADMIXTURE EUR percentage closer to 95% in the previous section (more similar to the chromosome painting results) and was even less accurate by that measure. However, if you say that you can only consider individuals with 1 primary ancestry assignment with a >50% or >70% contribution (for me, EUR), then these results are all compatible.
On top of that, this is not really a fair comparison for ancestry, since the imputations were made using CEU, GBR, and ACB 1000 Genomes reference samples (the set of 286 reference samples used for IBD/kinship self-recovery).
In contrast, these are the results if I only use the GIH population samples as the reference for the imputations, and kinship similar decreases a little (~0.457) and this is what the supervised ADMIXTURE assignments look like (which introduces the expected bias based upon the reference population):
The run-time was also noticeably shorter when I used 1 population for imputation (versus 3 populations for imputation).
That said, if you had a more diverse set of samples to test, then that should also be taken into consideration. For example, if I guessed the wrong ancestry for imputation, then that can cause some noticeable problems.
While it looks like STITCH off-target read analysis might provide improvement on first impression, the need to know the ancestry for a sample in advance confounds this particular application. Namely, I would say this re-emphasizes that you need to be careful to read too much into the EAS versus AMR differences that I reported earlier, since I used the most relevant populations for myself and those results are therefore not completely unbiased (think of it like the opposite of the SAS/GIH imputation test).
I ended up using considerably fewer off-target genotypes than I was expecting (>50,000 bp off-target sequence), but you can see that I did need to use more sequence (closer to the target regions) for the GLIMPSE analysis below.
Strategy #2b: Use Off-Target Reads for GLIMPSE lcWGS Analysis
In the lcWGS self-identification for my own sample, the performance of GLIMPSE was lower than STITCH, but the run-time was noticeably faster and all the 1000 Genome samples were used (so, it was not designed to use a subset of most related populations to improve performance).
Because the run-time was shorter, I tested multiple flanking distances from the coding regions. This was important because I needed to use sequence closer to the target regions to get self-identification more similar to STITCH:
| Sample 1 | Sample 2 | Kinship |
| GLIMPSE (Genos Off- Target) 50,000 bp flanking | 23andMe SNP chip | 0.362039 |
| GLIMPSE (Genos Off- Target) 10,000 bp flanking | 23andMe SNP chip | 0.449494 |
| GLIMPSE (Genos Off- Target) 2,000 bp flanking | 23andMe SNP chip | 0.475468 |
In all 3 cases above, the number of genotypes imputed by GLIMPSE was identical (91,296 variants). Unlike STITCH, there was no "PASS" filter for these variants. However, if this means that the variants were more accurate when more reads were used, then I believe the number of variants that would have had a "PASS" status should have also increased.
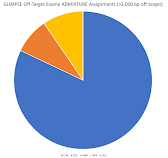
If you use the >10,000 bp and >2,000 bp flanking sequence, then you can see my ancestry results below (respectively):
I am not sure if it matters that I had to use sequence closer to the target (coding) regions, but these look similar to the regular on-target variant ancestry results (which were not as computationally intenstive and took up less storage space).
You can also see some more details about the GLIMPSE analysis on my samples here.
Finally, this was my first attempt at estimating ancestry from my Exome samples, so I think that there is likely some room for improvement. Nevertheless, I hope this is useful as a starting point for discussion (for what I thought was more-or-less the most straightforward application of existing methods).
Disclaimer: I think it is important to emphasize that this amount work may not be enough to say the strategy is completely free of errors, and it is also not enough to say the process should be scaled up for more samples.
However, if this post and the public GitHub code can play a similar role as a public lab notebook like the LOG.txt file for the RNA-Seq differential expression limits test, then that might help with communicating effort (and/or a partial contribution to a process, for both positive and negative results).
Change Log:
7/3/2020 - public post date
7/4/2020 - minor changes (including clarification of summary/discussion)
7/5/2020 - minor changes (including adding link to initialized GLIMPSE GitHub subfolder)
7/6/2020 - minor changes
7/4/2020 - minor changes (including clarification of summary/discussion)
7/5/2020 - minor changes (including adding link to initialized GLIMPSE GitHub subfolder)
7/6/2020 - minor changes
8/1/2020 - add GLIMPSE results